Agrobacterium tumefaciens
| Agrobacterium tumefaciens | |
|---|---|
 | |
| Agrobacterium tumefaciens attaching itself to a carrot cell | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Pseudomonadota |
| Class: | Alphaproteobacteria |
| Order: | Hyphomicrobiales |
| Family: | Rhizobiaceae |
| Genus: | Agrobacterium |
| Species: | A. tumefaciens |
| Binomial name | |
| Agrobacterium tumefaciens (Smith and Townsend 1907) Conn 1942 (Approved Lists 1980) | |
| Type strain | |
| ATCC 4720[1][2][a] | |
| Synonyms[7][8][2] | |
| Homotypic synonyms
Heterotypic synonyms Agrobacterium radiobacter (Beijerinck and van Delden 1902) Conn 1942 (Approved Lists 1980) is NOT a synonym.[2] The two used to be synonimized[6] on the basis of an unjustified type strain change in the Approved Lists of 1980, now reverted.[2] | |
Agrobacterium tumefaciens[3][2] (also known as Rhizobium radiobacter) is the causal agent of crown gall disease (the formation of tumours) in over 140 species of eudicots. It is a rod-shaped, Gram-negative soil bacterium.[4] Symptoms are caused by the insertion of a small segment of DNA (known as T-DNA, for 'transfer DNA', not to be confused with tRNA that transfers amino acids during protein synthesis), from a plasmid into the plant cell,[9] which is incorporated at a semi-random location into the plant genome. Plant genomes can be engineered by use of Agrobacterium for the delivery of sequences hosted in T-DNA binary vectors.
Agrobacterium tumefaciens is an Alphaproteobacterium of the family Rhizobiaceae, which includes the nitrogen-fixing legume symbionts. Unlike the nitrogen-fixing symbionts, tumor-producing Agrobacterium species are pathogenic and do not benefit the plant. The wide variety of plants affected by Agrobacterium makes it of great concern to the agriculture industry.[10]
Economically, A. tumefaciens is a serious pathogen of walnuts, grape vines, stone fruits, nut trees, sugar beets, horse radish, and rhubarb, and the persistent nature of the tumors or galls caused by the disease make it particularly harmful for perennial crops.[11]
Agrobacterium tumefaciens grows optimally at 28 °C (82 °F). The doubling time can range from 2.5–4h depending on the media, culture format, and level of aeration.[12] At temperatures above 30 °C (86 °F), A. tumefaciens begins to experience heat shock which is likely to result in errors in cell division.[12]
Conjugation
[edit]To be virulent, the bacterium contains a tumour-inducing plasmid (Ti plasmid or pTi) 200 kbp long, which contains the T-DNA and all the genes necessary to transfer it to the plant cell.[13] Many strains of A. tumefaciens do not contain a pTi.
Since the Ti plasmid is essential to cause disease, prepenetration events in the rhizosphere occur to promote bacterial conjugation - exchange of plasmids amongst bacteria. In the presence of opines, A. tumefaciens produces a diffusible conjugation signal called N-(3-oxo-octanoyl)-L-homoserine lactone (3OC8HSL) or the Agrobacterium autoinducer.[14] This activates the transcription factor TraR, positively regulating the transcription of genes required for conjugation.[15]
Infection methods
[edit]Agrobacterium tumefaciens infects the plant through its Ti plasmid. The Ti plasmid integrates a segment of its DNA, known as T-DNA, into the chromosomal DNA of its host plant cells. A. tumefaciens has flagella that allow it to swim through the soil towards photoassimilates that accumulate in the rhizosphere around roots. Some strains may chemotactically move towards chemical exudates from plants, such as acetosyringone and sugars, which indicate the presence of a wound in the plant through which the bacteria may enter. Phenolic compounds are recognised by the VirA protein, a transmembrane protein encoded in the virA gene on the Ti plasmid. Sugars are recognised by the chvE protein, a chromosomal gene-encoded protein located in the periplasmic space.[16]
At least 25 vir genes on the Ti plasmid are necessary for tumor induction.[17] In addition to their perception role, virA and chvE induce other vir genes. The VirA protein has autokinase activity: it phosphorylates itself on a histidine residue. Then the VirA protein phosphorylates the VirG protein on its aspartate residue. The virG protein is a cytoplasmic protein produced from the virG Ti plasmid gene. It is a transcription factor, inducing the transcription of the vir operons. The ChvE protein regulates the second mechanism of the vir genes' activation. It increases VirA protein sensitivity to phenolic compounds.[16]
Attachment is a two-step process. Following an initial weak and reversible attachment, the bacteria synthesize cellulose fibrils that anchor them to the wounded plant cell to which they were attracted. Four main genes are involved in this process: chvA, chvB, pscA, and att. The products of the first three genes apparently are involved in the actual synthesis of the cellulose fibrils. These fibrils also anchor the bacteria to each other, helping to form a microcolony.[citation needed]
VirC, the most important virulent protein, is a necessary step in the recombination of illegitimate recolonization. It selects the section of the DNA in the host plant that will be replaced and it cuts into this strand of DNA.[citation needed]
After production of cellulose fibrils, a calcium-dependent outer membrane protein called rhicadhesin is produced, which also aids in sticking the bacteria to the cell wall. Homologues of this protein can be found in other rhizobia. Currently, there are several reports on standardisation of protocol for the Agrobacterium-mediated transformation. The effect of different parameters such as infection time, acetosyringone, DTT, and cysteine have been studied in soybean (Glycine max).[18]
Possible plant compounds that initiate Agrobacterium to infect plant cells:[19]
- Acetosyringone and other phenolic compounds
- alpha-Hydroxyacetosyringone
- Catechol
- Ferulic acid
- Gallic acid
- p-Hydroxybenzoic acid
- Protocatechuic acid
- Pyrogallic acid
- Resorcylic acid
- Sinapinic acid
- Syringic acid
- Vanillin
Formation of the T-pilus
[edit]To transfer T-DNA into a plant cell, A. tumefaciens uses a type IV secretion mechanism, involving the production of a T-pilus. When acetosyringone and other substances are detected, a signal transduction event activates the expression of 11 genes within the VirB operon which are responsible for the formation of the T-pilus.
The pro-pilin is formed first. This is a polypeptide of 121 amino acids which requires processing by the removal of 47 residues to form a T-pilus subunit. The subunit was thought to be circularized by the formation of a peptide bond between the two ends of the polypeptide. However, high-resolution structure of the T-pilus revealed no cyclization of the pilin, with the overall organization of the pilin subunits being highly similar to those of other conjugative pili, such as F-pilus.[20]
Products of the other VirB genes are used to transfer the subunits across the plasma membrane. Yeast two-hybrid studies provide evidence that VirB6, VirB7, VirB8, VirB9 and VirB10 may all encode components of the transporter. An ATPase for the active transport of the subunits would also be required.
Transfer of T-DNA into the plant cell
[edit]
- Agrobacterium cell
- Agrobacterium chromosome
- Ti Plasmid (a. T-DNA, b. vir genes, c. replication origin, d. opines catabolism)
- Plant cell
- Plant mitochondria
- Plant chloroplast
- Plant nucleus
- VirA recognition
- VirA phosphorylates VirG
- VirG causes transcription of Vir genes
- Vir genes cut out T-DNA and form nucleoprotein complex ("T-complex")
- T-complex enters plant cytoplasm through T-pilus
- T-DNA enters into plant nucleus through nuclear pore
- T-DNA achieves integration
The T-DNA must be cut out of the circular plasmid. This is typically done by the Vir genes within the helper plasmid.[21] A VirD1/D2 complex nicks the DNA at the left and right border sequences. The VirD2 protein is covalently attached to the 5' end. VirD2 contains a motif that leads to the nucleoprotein complex being targeted to the type IV secretion system (T4SS). The structure of the T-pilus showed that the central channel of the pilus is too narrow to allow the transfer of the folded VirD2, suggesting that VirD2 must be partially unfolded during the conjugation process.[20]
In the cytoplasm of the recipient cell, the T-DNA complex becomes coated with VirE2 proteins, which are exported through the T4SS independently from the T-DNA complex. Nuclear localization signals, or NLSs, located on the VirE2 and VirD2, are recognised by the importin alpha protein, which then associates with importin beta and the nuclear pore complex to transfer the T-DNA into the nucleus. VIP1 also appears to be an important protein in the process, possibly acting as an adapter to bring the VirE2 to the importin. Once inside the nucleus, VIP2 may target the T-DNA to areas of chromatin that are being actively transcribed, so that the T-DNA can integrate into the host genome.
Genes in the T-DNA
[edit]Hormones
[edit]To cause gall formation, the T-DNA encodes genes for the production of auxin or indole-3-acetic acid via the IAM pathway. This biosynthetic pathway is not used in many plants for the production of auxin, so it means the plant has no molecular means of regulating it and auxin will be produced constitutively. Genes for the production of cytokinins are also expressed. This stimulates cell proliferation and gall formation.
Opines
[edit]The T-DNA contains genes for encoding enzymes that cause the plant to create specialized amino acid derivatives which the bacteria can metabolize, called opines.[22] Opines are a class of chemicals that serve as a source of nitrogen for A. tumefaciens, but not for most other organisms. The specific type of opine produced by A. tumefaciens C58 infected plants is nopaline.[23]
Two nopaline type Ti plasmids, pTi-SAKURA and pTiC58, were fully sequenced. "A. fabrum" C58, the first fully sequenced pathovar, was first isolated from a cherry tree crown gall. The genome was simultaneously sequenced by Goodner et al.[24] and Wood et al.[25] in 2001. The genome of A. tumefaciens C58 consists of a circular chromosome, two plasmids, and a linear chromosome. The presence of a covalently bonded circular chromosome is common to Bacteria, with few exceptions. However, the presence of both a single circular chromosome and single linear chromosome is unique to a group in this genus. The two plasmids are pTiC58, responsible for the processes involved in virulence, and pAtC58,[b] once dubbed the "cryptic" plasmid.[24][25]
The pAtC58 plasmid has been shown to be involved in the metabolism of opines and to conjugate with other bacteria in the absence of the pTiC58 plasmid.[26] If the Ti plasmid is removed, the tumor growth that is the means of classifying this species of bacteria does not occur.
Biotechnological uses
[edit]
The Asilomar Conference in 1975 established widespread agreement that recombinant techniques were insufficiently understood and needed to be tightly controlled.[27][28] The DNA transmission capabilities of Agrobacterium have been vastly explored in biotechnology as a means of inserting foreign genes into plants. Shortly after the Asilomar Conference, Marc Van Montagu and Jeff Schell discovered the gene transfer mechanism between Agrobacterium and plants, which resulted in the development of methods to alter the bacterium into an efficient delivery system for genetic engineering in plants.[29] The plasmid T-DNA that is transferred to the plant is an ideal vehicle for genetic engineering.[30] This is done by cloning a desired gene sequence into T-DNA binary vectors that will be used to deliver a sequence of interest into eukaryotic cells. This process has been performed using the firefly luciferase gene to produce glowing plants.[31] This luminescence has been a useful device in the study of plant chloroplast function and as a reporter gene.[31] It is also possible to transform Arabidopsis thaliana by dipping flowers into a broth of Agrobacterium: the seed produced will be transgenic. Under laboratory conditions, T-DNA has also been transferred to human cells, demonstrating the diversity of insertion application.[32]
The mechanism by which Agrobacterium inserts materials into the host cell is by a type IV secretion system which is very similar to mechanisms used by pathogens to insert materials (usually proteins) into human cells by type III secretion. It also employs a type of signaling conserved in many Gram-negative bacteria called quorum sensing.[citation needed] This makes Agrobacterium an important topic of medical research, as well.[citation needed]
Natural genetic transformation
[edit]Natural genetic transformation in bacteria is a sexual process involving the transfer of DNA from one cell to another through the intervening medium, and the integration of the donor sequence into the recipient genome by homologous recombination. A. tumefaciens can undergo natural transformation in soil without any specific physical or chemical treatment.[33]
Disease cycle
[edit]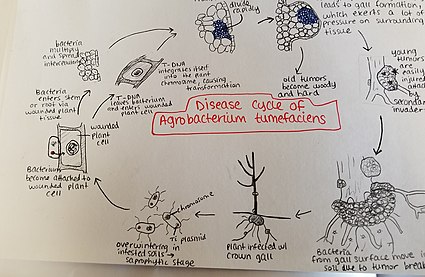
Agrobacterium tumefaciens overwinters in infested soils. Agrobacterium species live predominantly saprophytic lifestyles, so its common even for plant-parasitic species of this genus to survive in the soil for lengthy periods of time, even without host plant presence.[34] When there is a host plant present, however, the bacteria enter the plant tissue via recent wounds or natural openings of roots or stems near the ground. These wounds may be caused by cultural practices, grafting, insects, etc. Once the bacteria have entered the plant, they occur intercellularly and stimulate surrounding tissue to proliferate due to cell transformation. Agrobacterium performs this control by inserting the plasmid T-DNA into the plant's genome. See above for more details about the process of plasmid DNA insertion into the host genome. Excess growth of the plant tissue leads to gall formation on the stem and roots. These tumors exert significant pressure on the surrounding plant tissue, which causes this tissue to become crushed and/or distorted. The crushed vessels lead to reduced water flow in the xylem. Young tumors are soft and therefore vulnerable to secondary invasion by insects and saprophytic microorganisms. This secondary invasion causes the breakdown of the peripheral cell layers as well as tumor discoloration due to decay. Breakdown of the soft tissue leads to release of the Agrobacterium tumefaciens into the soil allowing it to restart the disease process with a new host plant.[35]
Disease management
[edit]Crown gall disease caused by Agrobacterium tumefaciens can be controlled by using various methods. The best way to control this disease is to take preventative measures, such as sterilizing pruning tools so as to avoid infecting new plants. Performing mandatory inspections of nursery stock and rejecting infected plants as well as not planting susceptible plants in infected fields are also valuable practices. Avoiding wounding the crowns/roots of the plants during cultivation is important for preventing disease. In horticultural techniques in which multiple plants are joined to grow as one, such as budding and grafting[36] these techniques lead to plant wounds. Wounds are the primary location of bacterial entry into the host plant. Therefore, it is advisable to perform these techniques during times of the year when Agrobacteria are not active. Control of root-chewing insects is also helpful to reduce levels of infection, since these insects cause wounds (aka bacterial entryways) in the plant roots.[35] It is recommended that infected plant material be burned rather than placed in a compost pile due to the bacteria's ability to live in the soil for many years.[37]
Biological control methods are also utilized in managing this disease. During the 1970s and 1980s, a common practice for treating germinated seeds, seedlings, and rootstock was to soak them in a suspension of K84. K84 is a strain of Rhizobium rhizogenes[38] (formerly classified under A. radiobacter, but later reclassified) which is a species related to A. tumefaciens but is not pathogenic. K84 produces a bacteriocin (agrocin 84) which is an antibiotic specific against related bacteria, including A. tumefaciens. This method, which was successful at controlling the disease on a commercial scale, had the risk of K84 transferring its resistance gene to the pathogenic Agrobacteria. Thus, in the 1990s, a deletion mutant strain based on K84, known as K1026, was created. This strain is just as successful in controlling crown gall as K84 without the caveat of resistance gene transfer.[39][40]
Environment
[edit]
Host, environment, and pathogen are extremely important concepts in regards to plant pathology. Agrobacteria have the widest host range of any plant pathogen,[41] so the main factor to take into consideration in the case of crown gall is environment. There are various conditions and factors that make for a conducive environment for A. tumefaciens when infecting its various hosts. The bacterium can't penetrate the host plant without an entry point such as a wound. Factors leading to wounds in plants include cultural practices, grafting, freezing injury, growth cracks, soil insects, and other animals in the environment causing damage to the plant. Consequently, in exceptionally harsh winters, it is common to have an increased incidence of crown gall due to the weather-related damage.[42] Along with this, there are methods of mediating infection of the host plant. For example, nematodes can act as a vector to introduce Agrobacterium into plant roots. More specifically, the root parasitic nematodes damage the plant cell, creating a wound for the bacteria to enter through.[43] Finally, temperature is a factor when considering A. tumefaciens infection. The optimal temperature for crown gall formation due to this bacterium is 22 °C (72 °F) because of the thermosensitivity of T-DNA transfer. Tumor formation is significantly reduced at higher temperature conditions.[44]
See also
[edit]References
[edit]- ^ Velazquez E, Flores-Felix JD, Sanchez-Juanes F, Igual JM, Peix A (2020). "Strain ATCC 4720T is the authentic type strain of Agrobacterium tumefaciens, which is not a later heterotypic synonym of Agrobacterium radiobacter". Int J Syst Evol Microbiol. 70 (9): 5172–5176. doi:10.1099/ijsem.0.004443. PMID 32915125.
- ^ a b c d e Arahal DR, Bull CT, Busse HJ, Christensen H, Chuvochina M, Dedysh SN, Fournier PE, Konstantinidis KT, Parker CT, Rossello-Mora R, Ventosa A, Göker M (27 April 2023). "Judicial Opinions 123–127". International Journal of Systematic and Evolutionary Microbiology. 72 (12). doi:10.1099/ijsem.0.005708. hdl:10261/295959. PMID 36748499. [N.B.: Judicial Opinion 127 assigns the strain ATCC 4720 as the type strain of Agrobacterium tumefaciens.]
- ^ a b c "Taxonomy browser (Agrobacterium tumefaciens)". National Center for Biotechnology Information. Retrieved 7 January 2024.
- ^ a b Smith EF, Townsend CO (April 1907). "A Plant-Tumor of Bacterial Origin". Science. 25 (643): 671–3. Bibcode:1907Sci....25..671S. doi:10.1126/science.25.643.671. PMID 17746161.
- ^ Delamuta JR, Scherer AJ, Ribeiro RA, Hungria M (2020). "Genetic diversity of Agrobacterium species isolated from nodules of common bean and soybean in Brazil, Mexico, Ecuador and Mozambique, and description of the new species Agrobacterium fabacearum sp. nov". International Journal of Systematic and Evolutionary Microbiology. 70 (7): 4233–4244. doi:10.1099/ijsem.0.004278. ISSN 1466-5034.
- ^ Tindall BJ, et al. (Judicial Commission) (2014). "Judicial Opinion No. 94: Agrobacterium radiobacter (Beijerinck and van Delden 1902) Conn 1942 has priority over Agrobacterium tumefaciens (Smith and Townsend 1907) Conn 1942 when the two are treated as members of the same species based on the principle of priority and Rule 23a, Note 1 as applied to the corresponding specific epithets". Int J Syst Evol Microbiol. 64 (Pt 10): 3590–3592. doi:10.1099/ijs.0.069203-0. PMID 25288664.
- ^ Buchanan RE (1965). "Proposal for rejection of the generic name Polymonas Lieske 1928". International Bulletin of Bacteriological Nomenclature and Taxonomy. 15 (1): 43–44. doi:10.1099/00207713-15-1-43.
- ^ Sawada H, Ieki H, Oyaizu H, Matsumoto S (1993). "Proposal for rejection of Agrobacterium tumefaciens and revised descriptions for the genus Agrobacterium and for Agrobacterium radiobacter and Agrobacterium rhizogenes". Int J Syst Bacteriol. 43 (4): 694–702. doi:10.1099/00207713-43-4-694. PMID 8240952.
- ^ Chilton MD, Drummond MH, Merlo DJ, Sciaky D, Montoya AL, Gordon MP, Nester EW (June 1977). "Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis". Cell. 11 (2): 263–271. doi:10.1016/0092-8674(77)90043-5. ISSN 0092-8674. PMID 890735. S2CID 7533482.
- ^ Moore LW, Chilton WS, Canfield ML (January 1997). "Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors". Applied and Environmental Microbiology. 63 (1): 201–7. Bibcode:1997ApEnM..63..201M. doi:10.1128/AEM.63.1.201-207.1997. PMC 1389099. PMID 16535484.
- ^ "Crown Galls". Missouri Botanical Garden. Retrieved December 2, 2019.
- ^ a b Morton ER, Fuqua C (February 2012). "Laboratory maintenance of Agrobacterium". Current Protocols in Microbiology. Chapter 1: Unit3D.1. doi:10.1002/9780471729259.mc03d01s24. ISBN 978-0471729259. PMC 3350319. PMID 22307549.
- ^ Gordon JE, Christie PJ (December 2014). "The Agrobacterium Ti Plasmids". Microbiology Spectrum. 2 (6). doi:10.1128/microbiolspec.PLAS-0010-2013. PMC 4292801. PMID 25593788.
- ^ Oger P, Farrand SK (2002). "Two Opines Control Conjugal Transfer of an Agrobacterium Plasmid by Regulating Expression of Separate Copies of the Quorum-Sensing Activator Gene traR". Journal of Bacteriology. 184 (4): 1121–1131. doi:10.1128/jb.184.4.1121-1131.2002. ISSN 0021-9193. PMC 134798. PMID 11807073.
- ^ Zhang HB, Wang LH, Zhang LH (2002). "Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens". Proceedings of the National Academy of Sciences. 99 (7): 4638–4643. Bibcode:2002PNAS...99.4638Z. doi:10.1073/pnas.022056699. ISSN 0027-8424. PMC 123700. PMID 11930013.
- ^ a b Gelvin SB (March 2003). "Agrobacterium-mediated plant transformation: the biology behind the "gene-jockeying" tool". Microbiology and Molecular Biology Reviews. 67 (1): 16–37, table of contents. doi:10.1128/mmbr.67.1.16-37.2003. PMC 150518. PMID 12626681.
- ^ Winans SC (1992). "Two-way chemical signaling in Agrobacterium-plant interactions". Microbiological Reviews. 56 (1): 12–31. doi:10.1128/mr.56.1.12-31.1992. ISSN 0146-0749. PMC 372852. PMID 1579105.
- ^ Barate PL, Kumar RR, Waghmare SG, Pawar KR, Tabe RH (2018). "Effect of different parameters on Agrobacterium mediated transformation in Glycine max". International Journal of Advanced Biological Research. 8 (1): 99–105.
- ^ US patent 6483013, "Method for agrobacterium mediated transformation of cotton", published 2002-11-19, assigned to Bayer BioScience N.V. (BE)
- ^ a b Beltran LC, Cvirkaite-Krupovic V, Miller J, Wang F, Kreutzberger MA, Patkowski JB, Costa TR, Schouten S, Levental I, Conticello VP, Egelman EH, Krupovic M (2023-02-07). "Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery". Nature Communications. 14 (1): 666. Bibcode:2023NatCo..14..666B. doi:10.1038/s41467-023-36349-8. ISSN 2041-1723. PMC 9905601. PMID 36750723.
- ^ Kroemer T. "A Guide to T-DNA Binary Vectors in Plant Transformation". GoldBio. Retrieved January 9, 2024.
- ^ Zupan J, Muth TR, Draper O, Zambryski P (July 2000). "The transfer of DNA from agrobacterium tumefaciens into plants: a feast of fundamental insights". The Plant Journal. 23 (1): 11–28. doi:10.1046/j.1365-313x.2000.00808.x. PMID 10929098.
- ^ Escobar MA, Civerolo EL, Polito VS, Pinney KA, Dandekar AM (January 2003). "Characterization of oncogene-silenced transgenic plants: implications for Agrobacterium biology and post-transcriptional gene silencing". Molecular Plant Pathology. 4 (1): 57–65. doi:10.1046/j.1364-3703.2003.00148.x. ISSN 1464-6722. PMID 20569363.
- ^ a b Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, et al. (December 2001). "Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58". Science. 294 (5550): 2323–2328. Bibcode:2001Sci...294.2323G. doi:10.1126/science.1066803. ISSN 0036-8075. PMID 11743194. S2CID 86255214.
- ^ a b Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, et al. (December 2001). "The genome of the natural genetic engineer Agrobacterium tumefaciens C58". Science. 294 (5550): 2317–23. Bibcode:2001Sci...294.2317W. CiteSeerX 10.1.1.7.9501. doi:10.1126/science.1066804. ISSN 0036-8075. PMID 11743193. S2CID 2761564.
- ^ Vaudequin-Dransart V, Petit A, Chilton WS, Dessaux Y (1998). "The cryptic plasmid of Agrobacterium tumefaciens cointegrates with the Ti plasmid and cooperates for opine degradation". Molecular Plant-Microbe Interactions. 11 (7): 583–591. doi:10.1094/mpmi.1998.11.7.583.
- ^ Berg P, Baltimore D, Brenner S, Roblin RO, Singer MF (1975-06-06). "Asilomar Conference on Recombinant DNA Molecules". Science. 188 (4192): 991–994. Bibcode:1975Sci...188..991B. doi:10.1126/science.1056638. ISSN 0036-8075. PMID 1056638.
- ^ National Research Council, Board on Agriculture and Natural Resources, Committee on Genetically Modified Pest-Protected Plants (2000). Genetically Modified Pest-Protected Plants: Science and Regulation. Washington, D.C.: National Academies Press. pp. xxiii, 263. doi:10.17226/9795. ISBN 978-0-309-06930-4. OCLC 894124744. PMID 25032472.
- ^ Schell J, Van Montagu M (1977). "The Ti-Plasmid of Agrobacterium Tumefaciens, A Natural Vector for the Introduction of NIF Genes in Plants?". In Hollaender A, Burris RH, Day PR, Hardy RW (eds.). Genetic Engineering for Nitrogen Fixation. Basic Life Sciences. Vol. 9. pp. 159–79. doi:10.1007/978-1-4684-0880-5_12. ISBN 978-1-4684-0882-9. PMID 336023.
- ^ Zambryski P, Joos H, Genetello C, Leemans J, Montagu MV, Schell J (1983). "Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity". The EMBO Journal. 2 (12): 2143–50. doi:10.1002/j.1460-2075.1983.tb01715.x. PMC 555426. PMID 16453482.
- ^ a b Root M (1988). "Glow in the dark biotechnology". BioScience. 38 (11): 745–747. doi:10.2307/1310781. JSTOR 1310781.
- ^ Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, Citovsky V (February 2001). "Genetic transformation of HeLa cells by Agrobacterium". Proceedings of the National Academy of Sciences of the United States of America. 98 (4): 1871–6. Bibcode:2001PNAS...98.1871K. doi:10.1073/pnas.041327598. PMC 29349. PMID 11172043.
- ^ Demanèche S, Kay E, Gourbière F, Simonet P (June 2001). "Natural transformation of Pseudomonas fluorescens and Agrobacterium tumefaciens in soil". Applied and Environmental Microbiology. 67 (6): 2617–21. Bibcode:2001ApEnM..67.2617D. doi:10.1128/AEM.67.6.2617-2621.2001. PMC 92915. PMID 11375171.
- ^ Schroth MN, Weinhold AR, Mccain AH (March 1971). "Biology and Control of Agrobacterium tumefaciens". Hilgardia. 40 (15): 537–552. doi:10.3733/hilg.v40n15p537.
- ^ a b Agrios GN (2005). Plant pathology (5th ed.). Amsterdam: Elsevier Academic Press. doi:10.1016/C2009-0-02037-6. ISBN 9780120445653. OCLC 55488155.
- ^ Bilderback T, Bir RE, Ranney TG (June 30, 2014). "Grafting and Budding Nursery Crop Plants". NC State Extension Publications. Retrieved December 12, 2017.
- ^ Koetter R, Grabowski M. "Crown gall". University of Minnesota Extension. Archived from the original on October 16, 2017. Retrieved October 15, 2017.
- ^ "Taxonomy browser (Rhizobium rhizogenes K84)". www.ncbi.nlm.nih.gov.
- ^ Ryder MH, Jones DA (October 1, 1991). "Biological Control of Crown Gall Using Using Agrobacterium Strains K84 and K1026". Functional Plant Biology. 18 (5): 571–579. doi:10.1071/pp9910571.
- ^ Kim JG, Park BK, Kim SU, Choi D, Nahm BH, Moon JS, Reader JS, Farrand SK, Hwang I (6 June 2006). "Bases of biocontrol: Sequence predicts synthesis and mode of action of agrocin 84, the Trojan Horse antibiotic that controls crown gall". Proceedings of the National Academy of Sciences. 103 (23): 8846–8851. doi:10.1073/pnas.0602965103. PMC 1482666.
- ^ Ellis MA (Apr 15, 2016). "Bacterial Crown Gall of Fruit Crops". Ohioline. Ohio State University Extension. Retrieved October 20, 2017.
- ^ "Crown Gall – A Growing Concern in Vineyards". Penn State Extension. October 19, 2017. Archived from the original on October 20, 2017. Retrieved October 20, 2017.
- ^ Karimi M, Van Montagu M, Gheysen G (November 2000). "Nematodes as vectors to introduce Agrobacterium into plant roots". Molecular Plant Pathology. 1 (6): 383–7. doi:10.1046/j.1364-3703.2000.00043.x. PMID 20572986. S2CID 35932276.
- ^ Dillen W, De Clereq J, Kapila J, Van Montagu ZM, Angenon G (1997-12-01). "The effect of temperature on Agrobacterium tumefaciens-mediated gene transfer to plants". The Plant Journal. 12 (6): 1459–1463. doi:10.1046/j.1365-313x.1997.12061459.x.
Further reading
[edit]- Dickinson M (2003). Molecular Plant Pathology. BIOS Scientific Publishers.
- Lai EM, Kado CI (August 2000). "The T-pilus of Agrobacterium tumefaciens". Trends in Microbiology. 8 (8): 361–9. doi:10.1016/s0966-842x(00)01802-3. PMID 10920395.
- Ward DV, Zupan JR, Zambryski PC (January 2002). "Agrobacterium VirE2 gets the VIP1 treatment in plant nuclear import". Trends in Plant Science. 7 (1): 1–3. doi:10.1016/s1360-1385(01)02175-6. PMID 11804814.
- Webster J, Thomson J (1988). "Genetic Analysis of an Agrobacterium tumefaciens strain producing an agrocin active against biotype 3 Pathogen". Molecular and General Genetics. 214 (1): 142–147. doi:10.1007/BF00340192. S2CID 180063.
External links
[edit] Media related to Agrobacterium tumefaciens at Wikimedia Commons
Media related to Agrobacterium tumefaciens at Wikimedia Commons- "Agrobacterium fabrum" C58 Genome Page — as sequenced by Cereon Genomics/University of Richmond


 French
French Deutsch
Deutsch