Chromatin immunoprecipitation
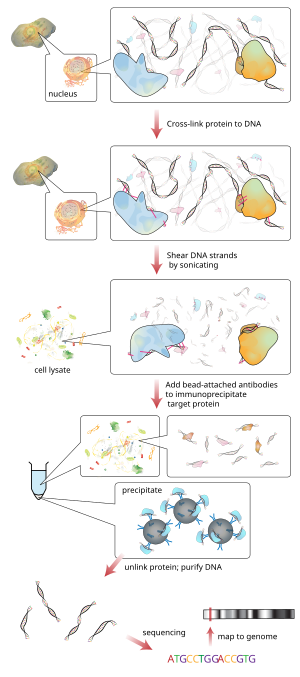
Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoters or other DNA binding sites, and possibly define cistromes. ChIP also aims to determine the specific location in the genome that various histone modifications are associated with, indicating the target of the histone modifiers.[1] ChIP is crucial for the advancements in the field of epigenomics and learning more about epigenetic phenomena.[2]
Briefly, the conventional method is as follows:
- DNA and associated proteins on chromatin in living cells or tissues are crosslinked (this step is omitted in Native ChIP).
- The DNA-protein complexes (chromatin-protein) are then sheared into ~500 bp DNA fragments by sonication or nuclease digestion.
- Cross-linked DNA fragments associated with the protein(s) of interest are selectively immunoprecipitated from the cell debris using an appropriate protein-specific antibody.
- The associated DNA fragments are purified and their sequence is determined. Enrichment of specific DNA sequences represents regions on the genome that the protein of interest is associated with in vivo.
Typical ChIP
[edit]There are mainly two types of ChIP, primarily differing in the starting chromatin preparation. The first uses reversibly cross-linked chromatin sheared by sonication called cross-linked ChIP (XChIP). Native ChIP (NChIP) uses native chromatin sheared by micrococcal nuclease digestion.[citation needed]
Cross-linked ChIP (XChIP)
[edit]Cross-linked ChIP is mainly suited for mapping the DNA target of transcription factors or other chromatin-associated proteins, and uses reversibly cross-linked chromatin as starting material. The agent for reversible cross-linking could be formaldehyde[3] or UV light.[4] Then the cross-linked chromatin is usually sheared by sonication, providing fragments of 300 - 1000 base pairs (bp) in length. Mild formaldehyde crosslinking followed by nuclease digestion has been used to shear the chromatin.[5] Chromatin fragments of 400 - 500bp have proven to be suitable for ChIP assays as they cover two to three nucleosomes.
Cell debris in the sheared lysate is then cleared by sedimentation and protein–DNA complexes are selectively immunoprecipitated using specific antibodies to the protein(s) of interest. The antibodies are commonly coupled to agarose, sepharose, or magnetic beads. Alternatively, chromatin-antibody complexes can be selectively retained and eluted by inert polymer discs.[6][7] The immunoprecipitated complexes (i.e., the bead–antibody–protein–target DNA sequence complex) are then collected and washed to remove non-specifically bound chromatin, the protein–DNA cross-link is reversed and proteins are removed by digestion with proteinase K. An epitope-tagged version of the protein of interest, or in vivo biotinylation [8] can be used instead of antibodies to the native protein of interest.
The DNA associated with the complex is then purified and identified by polymerase chain reaction (PCR), microarrays (ChIP-on-chip), molecular cloning and sequencing, or direct high-throughput sequencing (ChIP-Seq).[citation needed]
Native ChIP (NChIP)
[edit]Native ChIP is mainly suited for mapping the DNA target of histone modifiers. Generally, native chromatin is used as starting chromatin. As histones wrap around DNA to form nucleosomes, they are naturally linked. Then the chromatin is sheared by micrococcal nuclease digestion, which cuts DNA at the length of the linker, leaving nucleosomes intact and providing DNA fragments of one nucleosome (200bp) to five nucleosomes (1000bp) in length. Thereafter, methods similar to XChIP are used for clearing the cell debris, immunoprecipitating the protein of interest, removing protein from the immunoprecipitated complex, and purifying and analyzing the complex-associated DNA.[citation needed]
Comparison of XChIP and NChIP
[edit]The major advantage of NChIP is antibody specificity. Most antibodies to modified histones are raised against unfixed, synthetic peptide antigens. The epitopes they need to recognize in the XChIP may be disrupted or destroyed by formaldehyde cross-linking, particularly as the cross-links are likely to involve lysine e-amino groups in the N-terminals, disrupting the epitopes. This is likely to explain the consistently low efficiency of XChIP protocols compared to NChIP.
But XChIP and NChIP have different aims and advantages relative to each other. XChIP is for mapping target sites of transcription factors and other chromatin-associated proteins; NChIP is for mapping target sites of histone modifiers (see Table 1).
Comparison of ChIP-seq and ChIP-chip
[edit]Chromatin Immunoprecipitation sequencing, also known as ChIP-seq, is an experimental technique used to identify transcription factor binding events throughout an entire genome. Knowing how the proteins in the human body interact with DNA to regulate gene expression is a key component of our knowledge of human diseases and biological processes. ChIP-seq is the primary technique to complete this task, as it has proven to be extremely effective in resolving how proteins and transcription factors influence phenotypical mechanisms. Overall ChIP-seq has risen to be a very efficient method for determining these factors, but there is a rivaling method known as ChIP-on-chip.
ChIP-on-chip, also known as ChIP-chip, is an experimental technique used to isolate and identify genomic sites occupied by specific DNA-binding proteins in living cells. ChIP-on-chip is a relatively newer technique, as it was introduced in 2001 by Peggy Farnham and Michael Zhang. ChIP-on-chip gets its name by combining the methods of Chromatin Immunoprecipitation and DNA microarray, thus creating ChIP-on-chip.
The two methods seek similar results, as they both strive to find protein binding sites that can help identify elements in the human genome. Those elements in the human genome are important for the advancement of knowledge in human diseases and biological processes. The difference between ChIP-seq and ChIP-chip is established by the specific site of the protein binding identification. The main difference comes from the efficacy of the two techniques, ChIP-seq produces results with higher sensitivity and spatial resolution because of the wide range of genomic coverage. Even though ChIP-seq has proven to be more efficient than ChIP-chip, ChIP-seq is not always the first choice for scientists. The cost and accessibility of ChIP-seq is a major disadvantage, which has led to the more predominant use of ChIP-chip in laboratories across the world.[2]

Table 1 Advantages and disadvantages of NChIP and XChIP
| XChIP | NChIP | |
|---|---|---|
| Advantages | Suitable for transcriptional factors, or any other weakly binding chromatin associated proteins. Applicable to any organisms where native protein is hard to prepare | Testable antibody specificity Better antibody specificity as target protein naturally intact Better chromatin and protein revery efficiency due to better antibody specificity |
| Disadvantages | Inefficient chromatin recovery due to antibody target protein epitope disruption May cause false positive result due to fixation of transient proteins to chromatin Wide range of chromatin shearing size due to random cut by sonication. | Usually not suitable for non-histone proteins Nucleosomes may rearrange during digestion |
History and New ChIP methods
[edit]In 1984 John T. Lis and David Gilmour, at the time a graduate student in the Lis lab, used UV irradiation, a zero-length protein-nucleic acid crosslinking agent, to covalently cross-link proteins bound to DNA in living bacterial cells. Following lysis of cross-linked cells and immunoprecipitation of bacterial RNA polymerase, DNA associated with enriched RNA polymerase was hybridized to probes corresponding to different regions of known genes to determine the in vivo distribution and density of RNA polymerase at these genes. A year later they used the same methodology to study the distribution of eukaryotic RNA polymerase II on fruit fly heat shock genes. These reports are considered the pioneering studies in the field of chromatin immunoprecipitation.[9][10] XChIP was further modified and developed by Alexander Varshavsky and co-workers, who examined the distribution of histone H4 on heat shock genes using formaldehyde cross-linking.[11][12] This technique was extensively developed and refined thereafter.[13] NChIP approach was first described by Hebbes et al., 1988,[14] and has also been developed and refined quickly.[15] The typical ChIP assay usually takes 4–5 days and requires 106~ 107 cells at least. Now new techniques on ChIP could be achieved as few as 100~1000 cells and completed within one day.
- Bead-free ChIP: This novel method ChIP uses discs of inert, porous polymer functionalized with either Protein A or G in spin columns or microplates. The chromatin-antibody complex is selectively retained by the disc and eluted to obtain enriched DNA for downstream applications such as qPCR and sequencing. The porous environment is specifically designed to maximize capture efficiency and reduce non-specific binding. Due to less manual handling and optimized protocols, ChIP can be performed in 5 hours.[7]
- Carrier ChIP (CChIP): This approach could use as few as 100 cells by adding Drosophila cells as carrier chromatin to reduce loss and facilitate precipitation of the target chromatin. However, it demands highly specific primers for detection of the target cell chromatin from the foreign carrier chromatin background, and it takes two to three days.[16]
- Fast ChIP (qChIP): The fast ChIP assay reduced the time by shortening two steps in a typical ChIP assay: (i) an ultrasonic bath accelerates the rate of antibody binding to target proteins—and thereby reduces immunoprecipitation time (ii) a resin-based (Chelex-100) DNA isolation procedure reduces the time of cross-link reversal and DNA isolation. However, the fast protocol is suitable only for large cell samples (in the range of 106~107).[17][18] Up to 24 sheared chromatin samples can be processed to yield PCR-ready DNA in 5 hours, allowing multiple chromatin factors be probed simultaneously and/or looking at genomic events over several time points.[19]
- Quick and quantitative ChIP (Q2ChIP): The assay uses 100,000 cells as starting material and is suitable for up to 1,000 histone ChIPs or 100 transcription factor ChIPs. Thus many chromatin samples can be prepared in parallel and stored, and Q2ChIP can be undertaken in a day.[20]
- MicroChIP (μChIP): chromatin is usually prepared from 1,000 cells and up to 8 ChIPs can be done in parallel without carriers. The assay can also start with 100 cells, but only suit for one ChIP. It can also use small (1 mm3) tissue biopsies and microChIP can be done within one day.[21][22]
- Matrix ChIP: This is a microplate-based ChIP assay with increased throughput and a simplified procedure. All steps are done in microplate wells without sample transfers, enabling potential for automation. It enables 96 ChIP assays for histone and various DNA-bound proteins in a single day.[23]
- Pathology-ChIP (PAT-ChIP): This technique allows ChIP from pathology formalin-fixed and paraffin-embedded tissues and thus the use of pathology archives (even those that are several years old) for epigenetic analyses and the identification of candidate epigenetic biomarkers or targets.[24]
ChIP has also been applied for genome-wide analysis by combining with microarray technology (ChIP-on-chip) or second-generation DNA-sequencing technology (Chip-Sequencing). ChIP can also combine with paired-end tags sequencing in Chromatin Interaction Analysis using Paired End Tag sequencing (ChIA-PET), a technique developed for large-scale, de novo analysis of higher-order chromatin structures.[25][26][27]
Limitations
[edit]- Large Scale assays using ChIP is challenging using intact model organisms. This is because antibodies have to be generated for each TF, or, alternatively, transgenic model organisms expressing epitope-tagged TFs need to be produced.
- Researchers studying differential gene expression patterns in small organisms also face problems as genes expressed at low levels, in a small number of cells, in narrow time window.
- ChIP experiments cannot discriminate between different TF isoforms (Protein isoform).
See also
[edit]- ChIP-exo, a technique that adds exonuclease treatment to the ChIP process to obtain up to single base pair resolution of binding sites
- ChIP-on-chip, combines ChIP with microarray technology
- DamID, an alternative location mapping technique that does not require specific antibodies
- RIP-Chip, a similar technique to analyze RNA-protein interactions
References
[edit]- ^ Collas, Philippe. (January 2010). "The Current State of Chromatin Immunoprecipitation". Molecular Biotechnology. 45 (1): 87–100. doi:10.1007/s12033-009-9239-8. PMID 20077036. S2CID 24225210.
- ^ a b Rosenfeld, John M; Cooke, Tracy; Li, Zirong; Saito, Kan; Taganov, Konstantin; Thyagarajan, Bhaskar; Solache, Alejandra (March 2013). "Systematic optimization of parameters involved in preparation of chromatin and chromatin immunoprecipitation (ChIP) workflow". Epigenetics & Chromatin. 6 (S1): P122, 1756–8935–6-S1-P122. doi:10.1186/1756-8935-6-S1-P122. ISSN 1756-8935. PMC 3620580.
- ^ Jackson, Vaughn (November 1978). "Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent". Cell. 15 (3): 945–54. doi:10.1016/0092-8674(78)90278-7. PMID 569554. S2CID 25169609.
- ^ Gilmour DS, Lis JT (August 1985). "In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster". Molecular and Cellular Biology. 5 (8): 2009–18. doi:10.1128/mcb.5.8.2009. PMC 366919. PMID 3018544.
- ^ Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T (January 2002). "Methylation at arginine 17 of histone H3 is linked to gene activation". EMBO Reports. 3 (1): 39–44. doi:10.1093/embo-reports/kvf013. PMC 1083932. PMID 11751582.
- ^ Beynon, Amy L.; Parkes, Lindsay J.; Turner, Matthew L.; Knight, Steve; Conlan, Steve; Francis, Lewis; Stocks, Ben (September 2014). "Chromatrap 96: a new solid-state platform for high-throughput ChIP". Nature Methods. 11 (9): i–ii. doi:10.1038/nmeth.f.372. ISSN 1548-7091.
- ^ a b "Chromatrap".Revolutionary solid state platform for chromatin immunopreciptation.
- ^ Viens A; et al. (2004). "Use of protein biotinylation in vivo for chromatin immunoprecipitation". Analytical Biochemistry. 325 (1): 68–76. doi:10.1016/j.ab.2003.10.015. PMID 14715286.
- ^ Gilmour DS, Lis JT (1984). "Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes". Proc Natl Acad Sci U S A. 81 (14): 4275–9. Bibcode:1984PNAS...81.4275G. doi:10.1073/pnas.81.14.4275. PMC 345570. PMID 6379641.
- ^ Gilmour DS, Lis JT (August 1985). "In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster". Mol. Cell. Biol. 5 (8): 2009–18. doi:10.1128/mcb.5.8.2009. PMC 366919. PMID 3018544.
- ^ Varshavsky A (December 2008). "Discovery of cellular regulation by protein degradation". Journal of Biological Chemistry. 283 (50): 34469–89. doi:10.1074/jbc.X800009200. PMC 3259866. PMID 18708349.
- ^ Solomon, Mark J; Larsen Pamela L; Varshavsky, Alexander. (June 1988). "Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene". Cell. 53 (6): 937–47. doi:10.1016/S0092-8674(88)90469-2. PMID 2454748. S2CID 11169130.
- ^ Orlando V (March 2000). "Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation". Trends in Biochemical Sciences. 25 (3): 99–104. doi:10.1016/S0968-0004(99)01535-2. PMID 10694875.
- ^ Hebbes, Tim R; Thorne, Alan W; Crane-Robinson C (May 1988). "A direct link between core histone acetylation and transcriptionally active chromatin". The EMBO Journal. 7 (5): 1395–402. doi:10.1002/j.1460-2075.1988.tb02956.x. PMC 458389. PMID 3409869.
- ^ O'Neill, Laura P; Turner, Bryan M (September 2003). "Immunoprecipitation of native chromatin: NChIP". Methods. 31 (1): 76–82. doi:10.1016/S1046-2023(03)00090-2. PMID 12893176.
- ^ O'Neill, Laura P; VerMilyea, Matthew D; Turner, Bryan M (July 2006). "Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations". Nature Genetics. 38 (7): 835–41. doi:10.1038/ng1820. PMID 16767102. S2CID 28311996.
- ^ Nelson, Joel D; Denisenko, Oleg; Sova, Pavel; Bomsztyk, Karol (2006). "Fast chromatin immunoprecipitation assay". Nucleic Acids Research. 34 (1): e2. doi:10.1093/nar/gnj004. PMC 1325209. PMID 16397291.
- ^ Nelson, Joel D; Denisenko, Oleg; Bomsztyk, Karol (2006). "Protocol for the fast chromatin immunoprecipitation (ChIP) method". Nature Protocols. 1 (1): 179–85. doi:10.1038/nprot.2006.27. PMID 17406230. S2CID 20577722.
- ^ Nelson J, Denisenko O, Bomsztyk K (2009). "The Fast Chromatin Immunoprecipitation Method". Chromatin Immunoprecipitation Assays. Methods in Molecular Biology. Vol. 567. pp. 45–57. doi:10.1007/978-1-60327-414-2_3. ISBN 978-1-60327-413-5. PMID 19588084.
- ^ Dahl, John Arne; Collas, Philippe (April 2007). "Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells". Stem Cells. 25 (4): 1037–46. doi:10.1634/stemcells.2006-0430. PMID 17272500.
- ^ Dahl, John Arne; Collas, Philippe (2008). "A rapid micro chromatin immunoprecipitation assay (microChIP)". Nature Protocols. 3 (6): 1032–45. doi:10.1038/nprot.2008.68. PMID 18536650. S2CID 29529307.
- ^ Dahl, John Arne; Collas, Philippe (2009). "μChIP: Chromatin Immunoprecipitation for Small Cell Numbers". Chromatin Immunoprecipitation Assays. Methods in Molecular Biology. Vol. 567. pp. 59–74. doi:10.1007/978-1-60327-414-2_4. ISBN 978-1-60327-413-5. PMID 19588085.
- ^ Flanagin, Steve; Nelson, Joel D; Castner, David G; Denisenko, Oleg; Bomsztyk, Karol (February 2008). "Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events". Nucleic Acids Research. 36 (3): e17. doi:10.1093/nar/gkn001. PMC 2241906. PMID 18203739.
- ^ Fanelli, Mirco; Amatori, Stefano; Barozzi, Iros; Soncini, Matias; Zuffo, Roberto Dal; Bucci, Gabriele; Capra, Maria; Quarto, Micaela; Dellino, Gaetano Ivan (2010-12-14). "Pathology tissue–chromatin immunoprecipitation, coupled with high-throughput sequencing, allows the epigenetic profiling of patient samples". Proceedings of the National Academy of Sciences. 107 (50): 21535–21540. Bibcode:2010PNAS..10721535F. doi:10.1073/pnas.1007647107. ISSN 0027-8424. PMC 3003125. PMID 21106756.
- ^ Fullwood, Melissa J; Han, Yuyuan; Wei, Chia-Lin; Ruan, Xiaoan; Ruan, Yijun (January 2010). Chromatin interaction analysis using paired-end tag sequencing. Vol. Chapter 21. pp. Unit 21.15.1–25. doi:10.1002/0471142727.mb2115s89. ISBN 978-0471142720. PMC 6924956. PMID 20069536.
{{cite book}}:|journal=ignored (help) - ^ Li, Guoliang; Fullwood, Melissa J; Xu, Han; Mulawadi, Fabianus Hendriyan; Velkov, Stoyan; Vega, Vinsensius; Ariyaratne, Pramila Nuwantha; Mohamed, Yusoff Bin; Ooi, Hong-Sain; Tennakoon, Chandana; Wei, Chia-Lin; Ruan, Yijun; Sung, Wing-Kin (February 2010). "ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing". Genome Biology. 11 (2): R22. doi:10.1186/gb-2010-11-2-r22. PMC 2872882. PMID 20181287.
- ^ "ChIA-PET: Novel Method For 3-D Whole Genome Mapping Research". ScienceDaily. Agency for Science, Technology and Research (A*STAR), Singapore. 2009-11-08. Retrieved 14 March 2010.
External links
[edit]- Chromatin+immunoprecipitation at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- EpigenomeNOE.com
- Chromatin Immunopreciptation (ChIP) on Unfixed Chromatin from Cells and Tissues to Analyze Histone Modifications
- Chromatin Immunoprecipitation (ChIP) of Protein Complexes: Mapping of Genomic Targets of Nuclear Proteins in Cultured Cells


 French
French Deutsch
Deutsch