Hypervariable region
It has been suggested that this article should be split into articles titled Hypervariable region (nucleic acid) and Hypervariable region (antibody). (discuss) (June 2022) |
A hypervariable region (HVR) is a location within nuclear DNA or the D-loop of mitochondrial DNA in which base pairs of nucleotides repeat (in the case of nuclear DNA) or have substitutions (in the case of mitochondrial DNA). Changes or repeats in the hypervariable region are highly polymorphic.
Mitochondrial
[edit]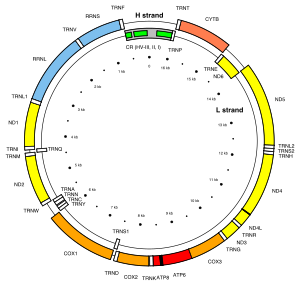
There are two mitochondrial hypervariable regions used in human mitochondrial genealogical DNA testing. HVR1 is considered a "low resolution" region and HVR2 is considered a "high resolution" region. Getting HVR1 and HVR2 DNA tests can help determine one's haplogroup. In the revised Cambridge Reference Sequence of the human mitogenome, the most variable sites of HVR1 are numbered 16024-16383 (this subsequence is called HVR-I), and the most variable sites of HVR2 are numbered 57-372 (i.e., HVR-II) and 438-574 (i.e., HVR-III).[1][2]
In some bony fishes, for example certain Protacanthopterygii and Gadidae, the mitochondrial control region evolves remarkably slowly. Even functional mitochondrial genes accumulate mutations faster and more freely. It is not known whether such hypovariable control regions are more widespread. In the Ayu (Plecoglossus altivelis), an East Asian protacanthopterygian, control region mutation rate is not markedly lowered, but sequence differences between subspecies are far lower in the control region than elsewhere. This phenomenon completely defies explanation at present.[3]
Antibodies
[edit]In antibodies, hypervariable regions form the antigen-binding site and are found on both light and heavy chains.[4] They also contribute to the specificity of each antibody.[4] In a variable domain, the 3 HV segments of each heavy or light chain fold together at the N-terminus to form an antigen binding pocket.[citation needed]
See also
[edit]- Cambridge Reference Sequence
- Genealogical DNA test
- Human mitochondrial DNA haplogroup
- mtDNA control region
References
[edit]- ^ van Oven M, Kayser M (February 2009). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation. 30 (2): E386–94. doi:10.1002/humu.20921. PMID 18853457.
- ^ PhyloTree mt. "Annotated mtDNA reference sequences: revised Cambridge Reference Sequence (rCRS)". Retrieved on 4 February 2016.
- ^ Takeshima, Hirohiko; Iguchi, Kei-ichiro & Nishida, Mutsumi (2005): Unexpected Ceiling of Genetic Differentiation in the Control Region of the Mitochondrial DNA between Different Subspecies of the Ayu Plecoglossus altivelis. Zool. Sci. 22(4): 401–410. doi:10.2108/zsj.22.401 (HTML abstract)
- ^ a b Michael Stein; Paul Zei; Gloria Hwang; Radhika Breaden (2000). Cracking The Boards: USMLE Step 1. The Princeton Review. ISBN 9780375761638. Retrieved 5 September 2011.
Antibodies are remarkably specific, thanks to hypervariable regions in both light and heavy chains. The hyperbariable regions for the antigen-binding site.
External links
[edit]- Hypervariable+region at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- DNA: Forensic and Legal Applications, Explanation of Hypervariable Regions


 French
French Deutsch
Deutsch