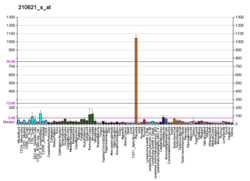
RAS p21 protein activator 1
RAS p21 protein activator 1 or RasGAP (Ras GTPase activating protein), also known as RASA1, is a 120-kDa cytosolic human protein that provides two principal activities:
- Inactivation of Ras from its active GTP-bound form to its inactive GDP-bound form by enhancing the endogenous GTPase activity of Ras, via its C-terminal GAP domain
- Mitogenic signal transmission towards downstream interacting partners through its N-terminal SH2-SH3-SH2 domains
The protein encoded by this gene is located in the cytoplasm and is part of the GAP1 family of GTPase-activating proteins. The gene product stimulates the GTPase activity of normal RAS p21 but not its oncogenic counterpart. Acting as a suppressor of RAS function, the protein enhances the weak intrinsic GTPase activity of RAS proteins resulting in the inactive GDP-bound form of RAS, thereby allowing control of cellular proliferation and differentiation. Mutations leading to changes in the binding sites of either protein are associated with basal cell carcinomas. Alternative splicing results in two isoforms where the shorter isoform, lacking the N-terminal hydrophobic region but retaining the same activity, appears to be abundantly expressed in placental but not adult tissues.[5]
Domains
[edit]RasGAP contains one SH3 domain and two SH2 domains, a PH domain, a C2 domain, and a GAP domain.
Interactions
[edit]RAS p21 protein activator 1 has been shown to interact with:
The mRNA can interact with Mir-132 microRNA; this process is linked to angiogenesis.[32]
Disease database
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000145715 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000021549 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Entrez Gene: RASA1 RAS p21 protein activator (GTPase activating protein) 1".
- ^ Chow A, Gawler D (October 1999). "Mapping the site of interaction between annexin VI and the p120GAP C2 domain". FEBS Lett. 460 (1): 166–72. doi:10.1016/s0014-5793(99)01336-8. PMID 10571081. S2CID 42114086.
- ^ Lee H, Park DS, Wang XB, Scherer PE, Schwartz PE, Lisanti MP (September 2002). "Src-induced phosphorylation of caveolin-2 on tyrosine 19. Phospho-caveolin-2 (Tyr(P)19) is localized near focal adhesions, remains associated with lipid rafts/caveolae, but no longer forms a high molecular mass hetero-oligomer with caveolin-1". J. Biol. Chem. 277 (37): 34556–67. doi:10.1074/jbc.M204367200. PMID 12091389.
- ^ Trentin GA, Yin X, Tahir S, Lhotak S, Farhang-Fallah J, Li Y, Rozakis-Adcock M (April 2001). "A mouse homologue of the Drosophila tumor suppressor l(2)tid gene defines a novel Ras GTPase-activating protein (RasGAP)-binding protein". J. Biol. Chem. 276 (16): 13087–95. doi:10.1074/jbc.M009267200. PMID 11116152.
- ^ Dunant NM, Wisniewski D, Strife A, Clarkson B, Resh MD (May 2000). "The phosphatidylinositol polyphosphate 5-phosphatase SHIP1 associates with the dok1 phosphoprotein in bcr-Abl transformed cells". Cell. Signal. 12 (5): 317–26. doi:10.1016/s0898-6568(00)00073-5. PMID 10822173.
- ^ Yamanashi Y, Baltimore D (January 1997). "Identification of the Abl- and rasGAP-associated 62 kDa protein as a docking protein, Dok". Cell. 88 (2): 205–11. doi:10.1016/s0092-8674(00)81841-3. PMID 9008161. S2CID 14205526.
- ^ Némorin JG, Duplay P (May 2000). "Evidence that Llck-mediated phosphorylation of p56dok and p62dok may play a role in CD2 signaling". J. Biol. Chem. 275 (19): 14590–7. doi:10.1074/jbc.275.19.14590. PMID 10799545.
- ^ Holland SJ, Gale NW, Gish GD, Roth RA, Songyang Z, Cantley LC, Henkemeyer M, Yancopoulos GD, Pawson T (July 1997). "Juxtamembrane tyrosine residues couple the Eph family receptor EphB2/Nuk to specific SH2 domain proteins in neuronal cells". EMBO J. 16 (13): 3877–88. doi:10.1093/emboj/16.13.3877. PMC 1170012. PMID 9233798.
- ^ Zisch AH, Pazzagli C, Freeman AL, Schneller M, Hadman M, Smith JW, Ruoslahti E, Pasquale EB (January 2000). "Replacing two conserved tyrosines of the EphB2 receptor with glutamic acid prevents binding of SH2 domains without abrogating kinase activity and biological responses". Oncogene. 19 (2): 177–87. doi:10.1038/sj.onc.1203304. PMID 10644995. S2CID 21872001.
- ^ Hock B, Böhme B, Karn T, Feller S, Rübsamen-Waigmann H, Strebhardt K (July 1998). "Tyrosine-614, the major autophosphorylation site of the receptor tyrosine kinase HEK2, functions as multi-docking site for SH2-domain mediated interactions". Oncogene. 17 (2): 255–60. doi:10.1038/sj.onc.1201907. PMID 9674711. S2CID 25714553.
- ^ Koehler JA, Moran MF (May 2001). "RACK1, a protein kinase C scaffolding protein, interacts with the PH domain of p120GAP". Biochem. Biophys. Res. Commun. 283 (4): 888–95. doi:10.1006/bbrc.2001.4889. PMID 11350068.
- ^ Briggs SD, Bryant SS, Jove R, Sanderson SD, Smithgall TE (June 1995). "The Ras GTPase-activating protein (GAP) is an SH3 domain-binding protein and substrate for the Src-related tyrosine kinase, Hck". J. Biol. Chem. 270 (24): 14718–24. doi:10.1074/jbc.270.24.14718. PMID 7782336.
- ^ a b Giglione C, Gonfloni S, Parmeggiani A (June 2001). "Differential actions of p60c-Src and Lck kinases on the Ras regulators p120-GAP and GDP/GTP exchange factor CDC25Mm". Eur. J. Biochem. 268 (11): 3275–83. doi:10.1046/j.1432-1327.2001.02230.x. PMID 11389730.
- ^ Molloy DP, Owen D, Grand RJ (July 1995). "Ras binding to a C-terminal region of GAP". FEBS Lett. 368 (2): 297–303. doi:10.1016/0014-5793(95)00657-u. PMID 7628625. S2CID 23151818.
- ^ Sprang SR (July 1997). "GAP into the breach". Science. 277 (5324): 329–30. doi:10.1126/science.277.5324.329. PMID 9518363. S2CID 22836050.
- ^ Liu YF, Deth RC, Devys D (March 1997). "SH3 domain-dependent association of huntingtin with epidermal growth factor receptor signaling complexes". J. Biol. Chem. 272 (13): 8121–4. doi:10.1074/jbc.272.13.8121. PMID 9079622.
- ^ Seely BL, Reichart DR, Staubs PA, Jhun BH, Hsu D, Maegawa H, Milarski KL, Saltiel AR, Olefsky JM (August 1995). "Localization of the insulin-like growth factor I receptor binding sites for the SH2 domain proteins p85, Syp, and GTPase activating protein". J. Biol. Chem. 270 (32): 19151–7. doi:10.1074/jbc.270.32.19151. PMID 7642582.
- ^ Sánchez-Margalet V, Najib S (October 2001). "Sam68 is a docking protein linking GAP and PI3K in insulin receptor signaling". Mol. Cell. Endocrinol. 183 (1–2): 113–21. doi:10.1016/s0303-7207(01)00587-1. PMID 11604231. S2CID 24594450.
- ^ Jabado N, Jauliac S, Pallier A, Bernard F, Fischer A, Hivroz C (September 1998). "Sam68 association with p120GAP in CD4+ T cells is dependent on CD4 molecule expression". J. Immunol. 161 (6): 2798–803. doi:10.4049/jimmunol.161.6.2798. PMID 9743338. S2CID 10463909.
- ^ Koch CA, Moran MF, Anderson D, Liu XQ, Mbamalu G, Pawson T (March 1992). "Multiple SH2-mediated interactions in v-src-transformed cells". Mol. Cell. Biol. 12 (3): 1366–74. doi:10.1128/mcb.12.3.1366. PMC 369570. PMID 1545818.
- ^ Ger M, Zitkus Z, Valius M (October 2011). "Adaptor protein Nck1 interacts with p120 Ras GTPase-activating protein and regulates its activity". Cell. Signal. 23 (10): 1651–8. doi:10.1016/j.cellsig.2011.05.019. PMID 21664272.
- ^ Farooqui T, Kelley T, Coggeshall KM, Rampersaud AA, Yates AJ (1999). "GM1 inhibits early signaling events mediated by PDGF receptor in cultured human glioma cells". Anticancer Res. 19 (6B): 5007–13. PMID 10697503.
- ^ Ekman S, Kallin A, Engström U, Heldin CH, Rönnstrand L (March 2002). "SHP-2 is involved in heterodimer specific loss of phosphorylation of Tyr771 in the PDGF beta-receptor". Oncogene. 21 (12): 1870–5. doi:10.1038/sj.onc.1205210. PMID 11896619. S2CID 35823546.
- ^ Chow A, Davis AJ, Gawler DJ (March 2000). "Identification of a novel protein complex containing annexin VI, Fyn, Pyk2, and the p120(GAP) C2 domain". FEBS Lett. 469 (1): 88–92. doi:10.1016/s0014-5793(00)01252-7. PMID 10708762. S2CID 21394463.
- ^ Zrihan-Licht S, Fu Y, Settleman J, Schinkmann K, Shaw L, Keydar I, Avraham S, Avraham H (March 2000). "RAFTK/Pyk2 tyrosine kinase mediates the association of p190 RhoGAP with RasGAP and is involved in breast cancer cell invasion". Oncogene. 19 (10): 1318–28. doi:10.1038/sj.onc.1203422. PMID 10713673.
- ^ Cacalano NA, Sanden D, Johnston JA (May 2001). "Tyrosine-phosphorylated SOCS-3 inhibits STAT activation but binds to p120 RasGAP and activates Ras". Nat. Cell Biol. 3 (5): 460–5. doi:10.1038/35074525. PMID 11331873. S2CID 19179597.
- ^ Brott BK, Decker S, O'Brien MC, Jove R (October 1991). "Molecular features of the viral and cellular Src kinases involved in interactions with the GTPase-activating protein". Mol. Cell. Biol. 11 (10): 5059–67. doi:10.1128/mcb.11.10.5059. PMC 361505. PMID 1717825.
- ^ Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, King PD, Weis SM, Cheresh DA (2010). "MicroRNA-132–mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis". Nat Med. 16 (8): 909–14. doi:10.1038/nm.2186. PMC 3094020. PMID 20676106.
Further reading
[edit]- Tocque B, Delumeau I, Parker F, et al. (1997). "Ras-GTPase activating protein (GAP): a putative effector for Ras". Cell. Signal. 9 (2): 153–8. doi:10.1016/S0898-6568(96)00135-0. PMID 9113414.
- Boon LM, Mulliken JB, Vikkula M (2005). "RASA1: variable phenotype with capillary and arteriovenous malformations". Curr. Opin. Genet. Dev. 15 (3): 265–9. doi:10.1016/j.gde.2005.03.004. PMID 15917201.


 French
French Deutsch
Deutsch